Plotting 23andMe markers density using karyoploteR
Learn how to plot 23andMe markers density on a karyogram.
Background
23andMe is a personal genomics and biotechnology company based in Mountain View, CA. It offers saliva-based direct to consumer genetic testing and it probably is one of the most popular choices among people who wish to learn more about their own genetic makeup, including genetic genealogy/ancestry analyses.
For genotyping, the company is using a technology known as DNA microarray, SNP array or SNP-chip. In short, SNP-chip is enabling us to “read” certain polymorphic sites in human DNA, the sites that are known to vary between individuals. Different versions of these polymorphic sites are, in turn, associated with several traits and features like eye color, blood group or a risk for developing certain disease. Now, it is not always the case that a certain version directly causes this or another phenotype. Often, a particular SNP is a good proxy for the phenotype, it tags the phenotype. This stems from the fact that we tend to inherit DNA in rather consecutive chunks (haplotypes or haploblocks) and often particular variant of a SNP is inherited with another variant in its neighborhood, the actual causative variant. Using this fact, one can save some money by genotyping only a limited number of SNPs to tell relatively much about many interesting traits. 23andMe is doing this. They have been using 3 different variants of their genotyping SNP-chip introduced in 2008, 2012 and 2014. I have been interested in visualising density of gentyped SNPs per fixed-length window along the genome to see how these 3 chips differ.
The data
23andMe uses Mendel Family as their showcase and we will look at the data generated for one of the members of the family – Lily Mendel (who happens to be a real person, Linda Avey, co-founder of the company). Data obtained with all three SNP-chips are available at SNPedia:
After unpacking the files, we see that the data format is rather simple – there are 4 columns describing each and every marker (SNP):
- rsid – unique rs identifier of the marker,
- chr – chromosome,
- pos – coordinates of the marker,
- gtype – the actual genotype, e.g. GG.
Analyses
Initial preparations
First, we have to load all the required packages and prepare our working environment. I have run itno some troubles with what turned out to be missing fonts. I have found a nice solution to this problem here.
#source("https://bioconductor.org/biocLite.R")
#biocLite("karyoploteR", )
library(extrafont)
# need only do this once!
# font_import(pattern="[A/a]rial", prompt=FALSE)
library("ggbio")
library("stringr")
library("karyoploteR")
library("biovizBase")Loading the data
Now, we can load the data and fix column names. Next, we convert the data into GRanges object required for plotting marker densities.
gdata <- read.table(file = '~/Downloads/genome_Lilly_Mendel_v4.txt',
comment.char = '#')
names(gdata) <- c('rsid', 'chr', 'pos', 'gtype')
regions <- GRanges(seqnames = paste0('chr', gdata$chr),
ranges = IRanges(start = gdata$pos, width = 2))Plotting markers density
Finally, there is time to plot markers density on a karyogram. A karyogram is an ideogram showing ordered chromosomes, a map of chromosomes. We plot markers density over each chromosome using kpPlotDensity. What, I think, is a nice feature in karyoploteR is that on the karyogram it shows so-called G-bands. Giemsa staining is a specific chemical reaction that stains different parts of chromosomes in different way. It is widely used in cytogenetics to visualise chromosomes and their structure. There is one important parameter here – the window size which I have empirically set to 1Mbp.
kp <- plotKaryotype(cex=.7) %>%
kpAddBaseNumbers(cex=.5) %>%
kpPlotDensity(data=regions,
window.size = 1e6,
col='slateblue',
border='slateblue')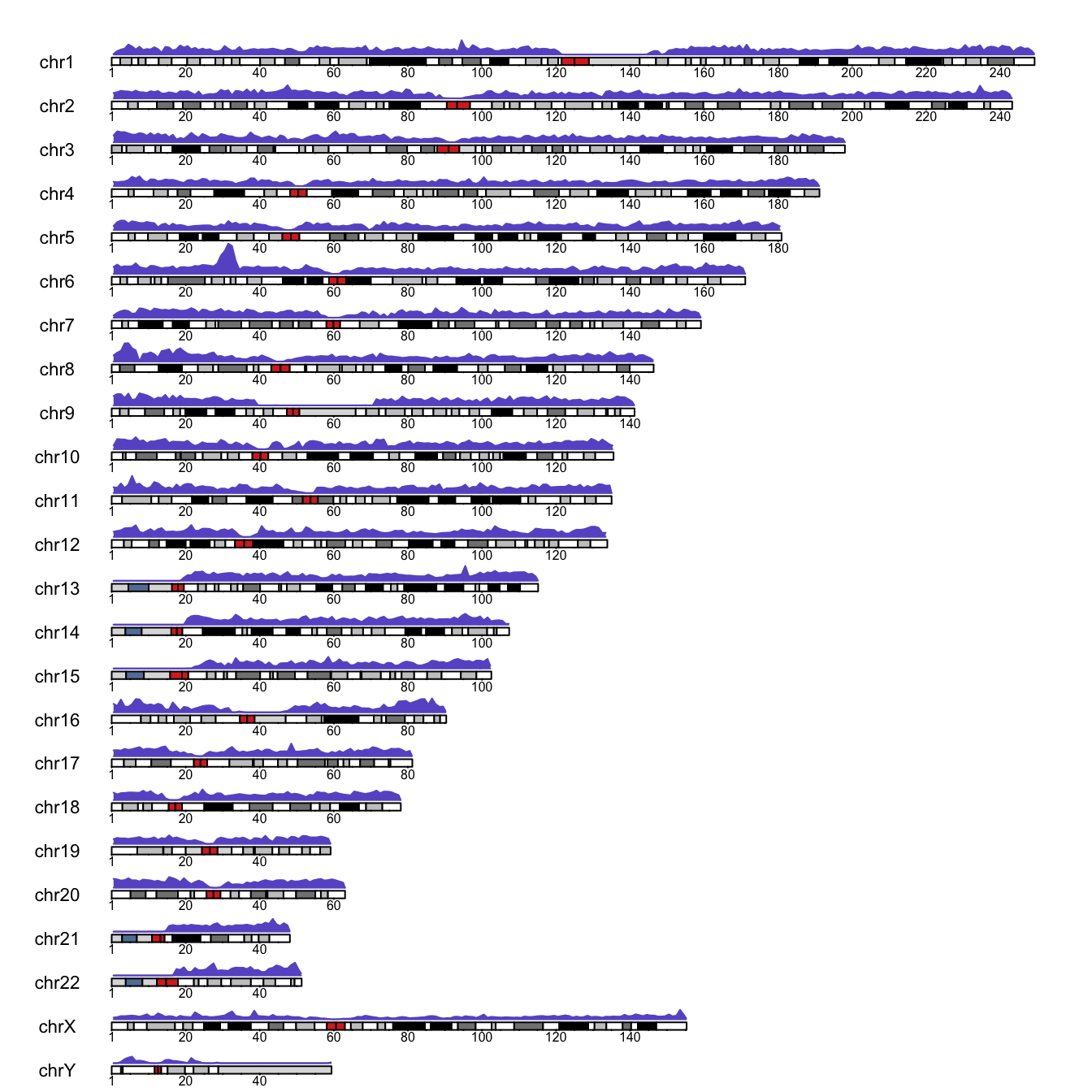
So what?
Well, I think the visualisation is pretty neat, although we are missing Y axis… We will fix this soon. Apart from this, we can see low markers density around centromeres (central parts of chromosomes G-stained in red). We also see a long stretch of no marker coverage on the Y chromosome. Perhaps this is the non-pseudoautosomal region of the chromosome. Similarly, the density is very low at the beginning of chromosomes 13, 14, 15, 21 and 22 and we have an exceptionally high density on the short arm of chromosome 6. I have no idea why and not much time to research right now…
Plotting the actual marker counts.
Ok, so this was the density. What if we want the actual counts per window? Easy enough!
counts <- kp$latest.plot$computed.values$density
data_counts <- GRanges(kp$latest.plot$computed.values$windows, y=counts)
max_count <- floor(max(counts) + .1 * max(counts))
# Change the default plotting parameters
# Values established in trial-and-error
pp <- getDefaultPlotParams(plot.type=1)
pp$leftmargin <- 0.15 # Avoid chr names to overlap with the plot
pp$topmargin <- 150 # Avoid overlap between chromosomes
pp$bottommargin <- 150
# Do the actual plotting
kp <- plotKaryotype(cex=.7, plot.params=pp) %>%
kpAddBaseNumbers(cex=.5) %>%
kpLines(data = data_counts, ymin = 0, ymax=max_count) %>%
kpAxis(cex=.5, ymin=0, ymax=max_count)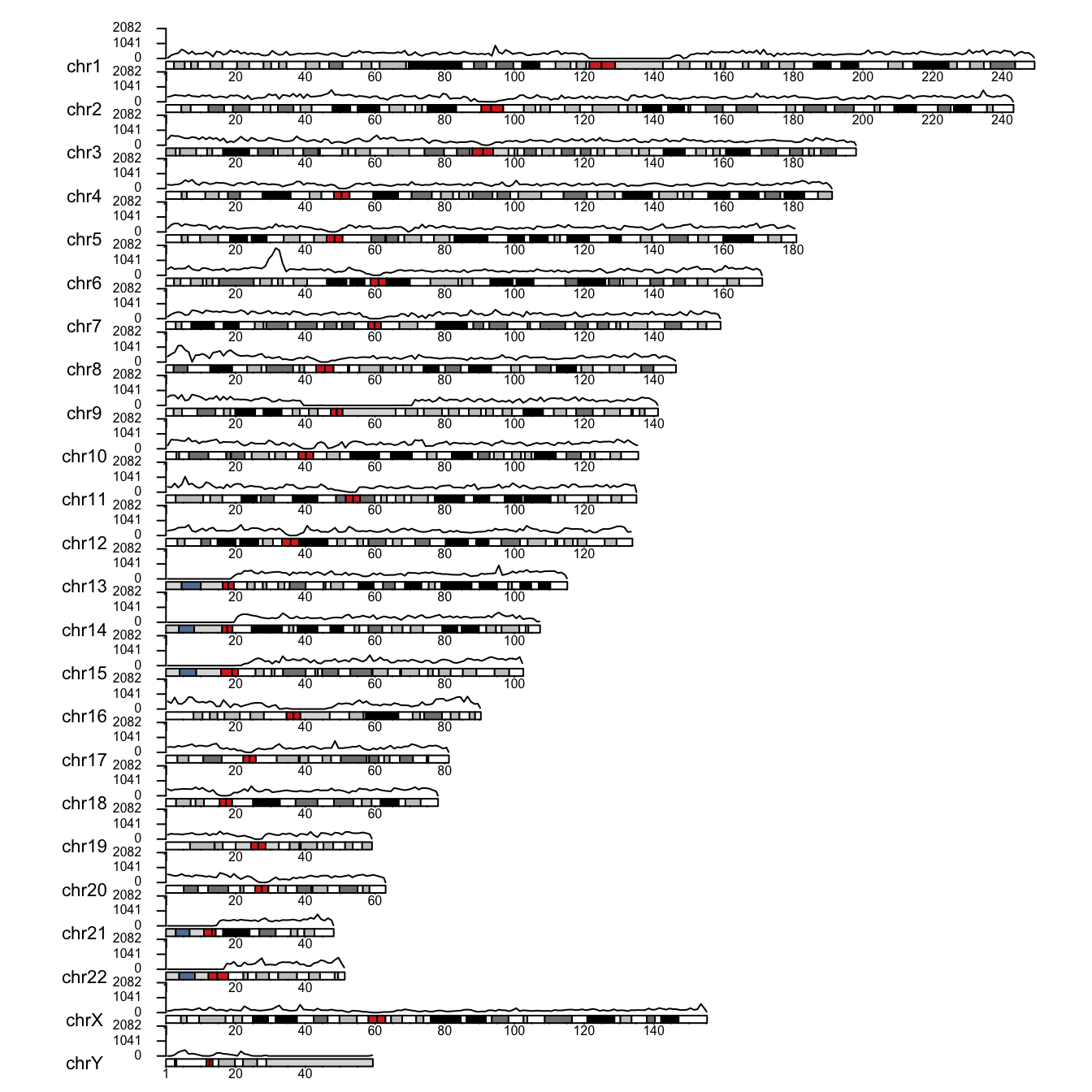
As you can see, I did not manage to completely avoid overlap between chromosomes and their data panels. This can probably be further refined by playing with graphical parameters though. You have got the idea, anyway.
Zooming in
We have previously spotted a region of high marker density on the short arm of chromosome 6. Let’s try to have a closer look.
pp <- getDefaultPlotParams(plot.type=1)
pp$leftmargin <- 0.15
pp$topmargin <- 120
to_zoom <- GRanges('chr6', IRanges(start=20e6, end=60e6))
kp <- plotKaryotype(cex=.7, plot.params=pp, zoom = to_zoom) %>%
kpAddBaseNumbers(cex=.5) %>%
kpAxis(cex=.5, ymin=0) %>%
kpBars(data=data_counts, y1 = data_counts$y, ymax=max_count)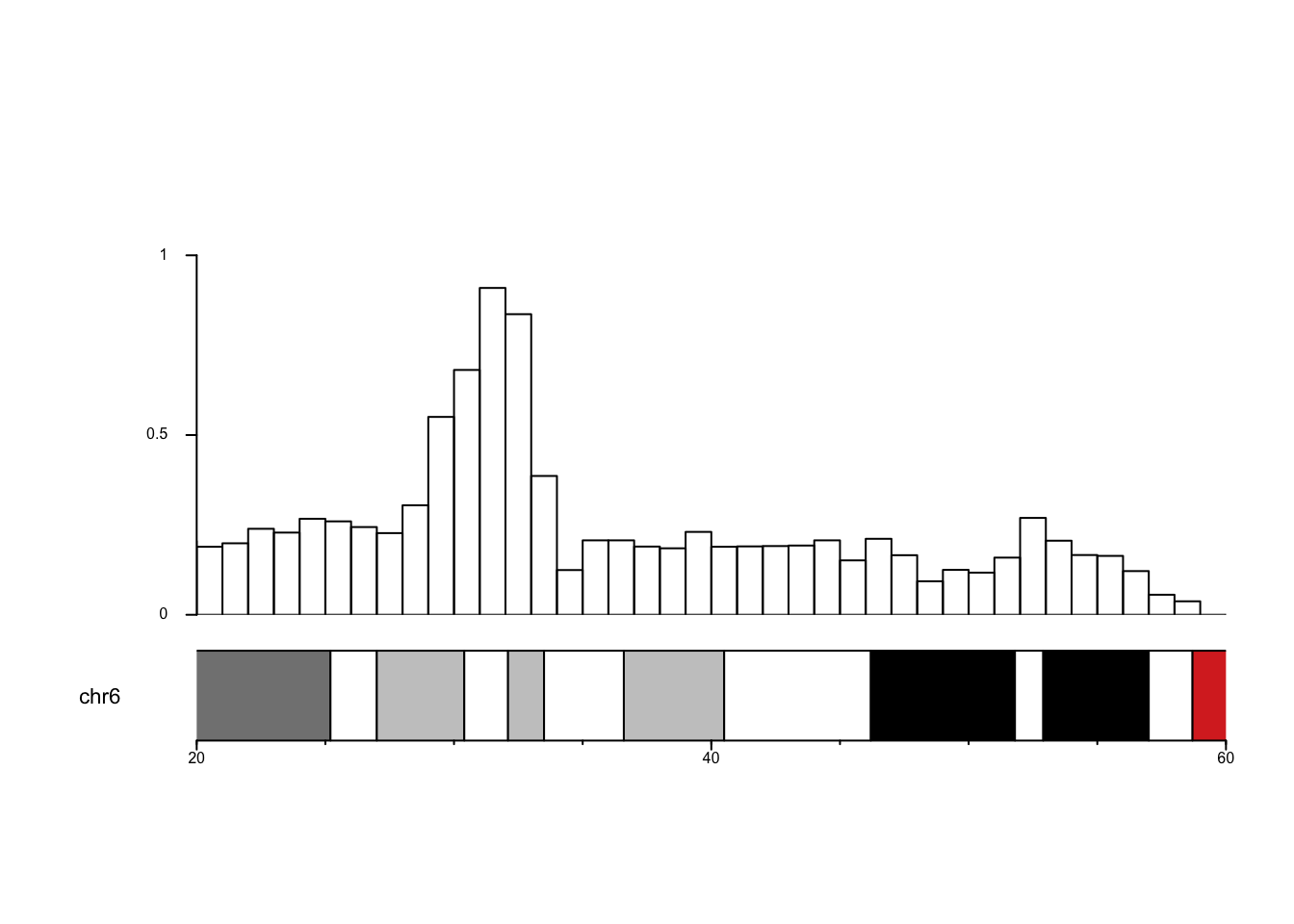
Well, almost perfect, only the Y axis has disappeared in a mysterious fashion… Do not know whether it is my fault or a feature of karyoploteR. Anyway, now we can see that the highest counts occur roughly between 30-35MBp. It’d be interesting to get gene names in the region together with some annotation.
Attention! There are two additional things I should have done here if it was a true research study:
- check the exact coords of high density, nt just eyeballing 30-35Mbp,
- since 23andMe is using hg19 assembly coordinates and Ensembl database is using newer hg38 assembly, (note that hg19 is the immediate predecessor of hg38!), I should have mapped the coords from one to another using, e.g. liftOver. I actually did and the coords for the region of interest are actually close enough not to bother for the purpose of this post.
Hint To find out the names of attributes and filters available in BioMart, go here and once you are done setting them, click on XML.
# Load the neccessary libraries
library("biomaRt")
# Retrieve the data
ensembl <- useMart("ENSEMBL_MART_ENSEMBL", dataset="hsapiens_gene_ensembl")
genes <- getBM(attributes=c("ensembl_gene_id", "external_gene_name", "description", "phenotype_description"),
filter=c("chromosome_name","start","end"),
values = list(chromosome='6', start='30000000', end='35000000'),
mart= ensembl)## Cache found# Make a nice table
library("knitr")
library("kableExtra")
genes %>% kable("html") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) %>%
scroll_box(height = "500px")| ensembl_gene_id | external_gene_name | description | phenotype_description |
|---|---|---|---|
| ENSG00000204623 | ZNRD1ASP | zinc ribbon domain containing 1 antisense, pseudogene [Source:HGNC Symbol;Acc:HGNC:13924] | |
| ENSG00000204622 | HLA-J | major histocompatibility complex, class I, J (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4967] | |
| ENSG00000237669 | AL671277.2 | HLA complex group 4 pseudogene 3 | |
| ENSG00000232757 | ETF1P1 | eukaryotic translation termination factor 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:3478] | |
| ENSG00000278104 | AL669914.3 | ||
| ENSG00000275856 | AL669914.2 | ||
| ENSG00000066379 | ZNRD1 | zinc ribbon domain containing 1 [Source:HGNC Symbol;Acc:HGNC:13182] | |
| ENSG00000278773 | AL669914.4 | ||
| ENSG00000204619 | PPP1R11 | protein phosphatase 1 regulatory inhibitor subunit 11 [Source:HGNC Symbol;Acc:HGNC:9285] | |
| ENSG00000204618 | RNF39 | ring finger protein 39 [Source:HGNC Symbol;Acc:HGNC:18064] | |
| ENSG00000204616 | TRIM31 | tripartite motif containing 31 [Source:HGNC Symbol;Acc:HGNC:16289] | |
| ENSG00000231226 | TRIM31-AS1 | TRIM31 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39761] | |
| ENSG00000252228 | AL669914.1 | ||
| ENSG00000204614 | TRIM40 | tripartite motif containing 40 [Source:HGNC Symbol;Acc:HGNC:18736] | |
| ENSG00000204613 | TRIM10 | tripartite motif containing 10 [Source:HGNC Symbol;Acc:HGNC:10072] | |
| ENSG00000204610 | TRIM15 | tripartite motif containing 15 [Source:HGNC Symbol;Acc:HGNC:16284] | |
| ENSG00000234127 | TRIM26 | tripartite motif containing 26 [Source:HGNC Symbol;Acc:HGNC:12962] | |
| ENSG00000233892 | PAIP1P1 | poly(A) binding protein interacting protein 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:18240] | |
| ENSG00000270604 | HCG17 | HLA complex group 17 [Source:HGNC Symbol;Acc:HGNC:31339] | |
| ENSG00000236475 | TRIM26BP | tripartite motif containing 26B, pseudogene [Source:HGNC Symbol;Acc:HGNC:31338] | |
| ENSG00000243753 | HLA-L | major histocompatibility complex, class I, L (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4970] | |
| ENSG00000280128 | AL662795.2 | TEC | |
| ENSG00000231074 | HCG18 | HLA complex group 18 [Source:HGNC Symbol;Acc:HGNC:31337] | |
| ENSG00000204599 | TRIM39 | tripartite motif containing 39 [Source:HGNC Symbol;Acc:HGNC:10065] | |
| ENSG00000248167 | TRIM39-RPP21 | TRIM39-RPP21 readthrough [Source:HGNC Symbol;Acc:HGNC:38845] | |
| ENSG00000241370 | RPP21 | ribonuclease P/MRP subunit p21 [Source:HGNC Symbol;Acc:HGNC:21300] | |
| ENSG00000224372 | HLA-N | major histocompatibility complex, class I, N (pseudogene) [Source:HGNC Symbol;Acc:HGNC:19406] | |
| ENSG00000224486 | AL662795.1 | HLA complex group 19 pseudogene | |
| ENSG00000236405 | UBQLN1P1 | ubiquilin 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21633] | |
| ENSG00000226577 | MICC | MHC class I polypeptide-related sequence C (pseudogene) [Source:HGNC Symbol;Acc:HGNC:7092] | |
| ENSG00000286301 | AL662873.1 | novel transcript | |
| ENSG00000229068 | TMPOP1 | thymopoietin pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21364] | |
| ENSG00000224936 | SUCLA2P1 | succinate-CoA ligase ADP-forming beta subunit pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21632] | |
| ENSG00000236603 | RANP1 | RAN pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21631] | |
| ENSG00000204592 | HLA-E | major histocompatibility complex, class I, E [Source:HGNC Symbol;Acc:HGNC:4962] | |
| ENSG00000235781 | LINC02569 | long intergenic non-protein coding RNA 2569 [Source:HGNC Symbol;Acc:HGNC:39760] | |
| ENSG00000204590 | GNL1 | G protein nucleolar 1 (putative) [Source:HGNC Symbol;Acc:HGNC:4413] | |
| ENSG00000204576 | PRR3 | proline rich 3 [Source:HGNC Symbol;Acc:HGNC:21149] | |
| ENSG00000204574 | ABCF1 | ATP binding cassette subfamily F member 1 [Source:HGNC Symbol;Acc:HGNC:70] | |
| ENSG00000216101 | MIR877 | microRNA 877 [Source:HGNC Symbol;Acc:HGNC:33660] | |
| ENSG00000204569 | PPP1R10 | protein phosphatase 1 regulatory subunit 10 [Source:HGNC Symbol;Acc:HGNC:9284] | |
| ENSG00000204568 | MRPS18B | mitochondrial ribosomal protein S18B [Source:HGNC Symbol;Acc:HGNC:14516] | |
| ENSG00000137343 | ATAT1 | alpha tubulin acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:21186] | |
| ENSG00000228415 | PTMAP1 | prothymosin alpha pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:9624] | |
| ENSG00000204564 | C6orf136 | chromosome 6 open reading frame 136 [Source:HGNC Symbol;Acc:HGNC:21301] | |
| ENSG00000204560 | DHX16 | DEAH-box helicase 16 [Source:HGNC Symbol;Acc:HGNC:2739] | Intellectual Disability Central Nervous System anomalies and Seizures |
| ENSG00000146112 | PPP1R18 | protein phosphatase 1 regulatory subunit 18 [Source:HGNC Symbol;Acc:HGNC:29413] | |
| ENSG00000137404 | NRM | nurim [Source:HGNC Symbol;Acc:HGNC:8003] | |
| ENSG00000230449 | RPL7P4 | ribosomal protein L7 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:21634] | |
| ENSG00000137337 | MDC1 | mediator of DNA damage checkpoint 1 [Source:HGNC Symbol;Acc:HGNC:21163] | |
| ENSG00000224328 | MDC1-AS1 | MDC1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39764] | |
| ENSG00000196230 | TUBB | tubulin beta class I [Source:HGNC Symbol;Acc:HGNC:20778] | Multiple benign circumferential skin creases on limbs |
| ENSG00000196230 | TUBB | tubulin beta class I [Source:HGNC Symbol;Acc:HGNC:20778] | Cortical dysplasia complex with other brain malformations 6 |
| ENSG00000196230 | TUBB | tubulin beta class I [Source:HGNC Symbol;Acc:HGNC:20778] | Circumferential Skin Creases Kunze Type |
| ENSG00000196230 | TUBB | tubulin beta class I [Source:HGNC Symbol;Acc:HGNC:20778] | SKIN CREASES CONGENITAL SYMMETRIC CIRCUMFERENTIAL 1 |
| ENSG00000272540 | AL662797.2 | novel transcript, antisense to TUBB | |
| ENSG00000137312 | FLOT1 | flotillin 1 [Source:HGNC Symbol;Acc:HGNC:3757] | |
| ENSG00000201988 | AL662797.1 | ||
| ENSG00000272273 | IER3-AS1 | IER3 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:53629] | |
| ENSG00000137331 | IER3 | immediate early response 3 [Source:HGNC Symbol;Acc:HGNC:5392] | |
| ENSG00000228022 | HCG20 | HLA complex group 20 [Source:HGNC Symbol;Acc:HGNC:31334] | |
| ENSG00000263608 | RN7SL353P | RNA, 7SL, cytoplasmic 353, pseudogene [Source:HGNC Symbol;Acc:HGNC:46369] | |
| ENSG00000214894 | LINC00243 | long intergenic non-protein coding RNA 243 [Source:HGNC Symbol;Acc:HGNC:30956] | |
| ENSG00000237923 | LINC02570 | long intergenic non-protein coding RNA 2570 [Source:HGNC Symbol;Acc:HGNC:39766] | |
| ENSG00000202241 | RN7SKP186 | RN7SK pseudogene 186 [Source:HGNC Symbol;Acc:HGNC:45910] | |
| ENSG00000237775 | DDR1-DT | DDR1 divergent transcript [Source:HGNC Symbol;Acc:HGNC:28694] | |
| ENSG00000204580 | DDR1 | discoidin domain receptor tyrosine kinase 1 [Source:HGNC Symbol;Acc:HGNC:2730] | |
| ENSG00000284370 | MIR4640 | microRNA 4640 [Source:HGNC Symbol;Acc:HGNC:41561] | |
| ENSG00000213780 | GTF2H4 | general transcription factor IIH subunit 4 [Source:HGNC Symbol;Acc:HGNC:4658] | |
| ENSG00000288473 | AL669830.2 | novel protein | |
| ENSG00000137411 | VARS2 | valyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:21642] | Combined oxidative phosphorylation defect type 20 |
| ENSG00000137411 | VARS2 | valyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:21642] | Combined oxidative phosphorylation deficiency 20 |
| ENSG00000196260 | SFTA2 | surfactant associated 2 [Source:HGNC Symbol;Acc:HGNC:18386] | |
| ENSG00000252761 | AL669830.1 | ||
| ENSG00000168631 | MUCL3 | mucin like 3 [Source:HGNC Symbol;Acc:HGNC:21666] | Diffuse panbronchiolitis |
| ENSG00000233529 | HCG21 | HLA complex group 21 [Source:HGNC Symbol;Acc:HGNC:31335] | |
| ENSG00000275906 | NAPGP2 | N-ethylmaleimide-sensitive factor attachment protein, gamma pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:45166] | |
| ENSG00000204544 | MUC21 | mucin 21, cell surface associated [Source:HGNC Symbol;Acc:HGNC:21661] | |
| ENSG00000261272 | MUC22 | mucin 22 [Source:HGNC Symbol;Acc:HGNC:39755] | |
| ENSG00000228789 | HCG22 | HLA complex group 22 (gene/pseudogene) [Source:HGNC Symbol;Acc:HGNC:27780] | |
| ENSG00000222895 | RNU6-1133P | RNA, U6 small nuclear 1133, pseudogene [Source:HGNC Symbol;Acc:HGNC:48096] | |
| ENSG00000204542 | C6orf15 | chromosome 6 open reading frame 15 [Source:HGNC Symbol;Acc:HGNC:13927] | |
| ENSG00000204540 | PSORS1C1 | psoriasis susceptibility 1 candidate 1 [Source:HGNC Symbol;Acc:HGNC:17202] | |
| ENSG00000204539 | CDSN | corneodesmosin [Source:HGNC Symbol;Acc:HGNC:1802] | Hypotrichosis simplex of the scalp |
| ENSG00000204539 | CDSN | corneodesmosin [Source:HGNC Symbol;Acc:HGNC:1802] | Peeling skin syndrome type B |
| ENSG00000204539 | CDSN | corneodesmosin [Source:HGNC Symbol;Acc:HGNC:1802] | HYPOTRICHOSIS 2 |
| ENSG00000204539 | CDSN | corneodesmosin [Source:HGNC Symbol;Acc:HGNC:1802] | PEELING SKIN SYNDROME 1 |
| ENSG00000204538 | PSORS1C2 | psoriasis susceptibility 1 candidate 2 [Source:HGNC Symbol;Acc:HGNC:17199] | |
| ENSG00000238211 | POLR2LP1 | RNA polymerase II subunit L pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31340] | |
| ENSG00000204536 | CCHCR1 | coiled-coil alpha-helical rod protein 1 [Source:HGNC Symbol;Acc:HGNC:13930] | |
| ENSG00000137310 | TCF19 | transcription factor 19 [Source:HGNC Symbol;Acc:HGNC:11629] | |
| ENSG00000204531 | POU5F1 | POU class 5 homeobox 1 [Source:HGNC Symbol;Acc:HGNC:9221] | |
| ENSG00000204528 | PSORS1C3 | psoriasis susceptibility 1 candidate 3 [Source:HGNC Symbol;Acc:HGNC:17203] | |
| ENSG00000272501 | AL662844.4 | novel transcript, antisense to HCG27 | |
| ENSG00000206344 | HCG27 | HLA complex group 27 [Source:HGNC Symbol;Acc:HGNC:27366] | |
| ENSG00000271821 | AL662844.3 | novel transcript | |
| ENSG00000255726 | AL662844.1 | mitochondrial coiled-coil domain 1 (MCCD1) pseudogene | |
| ENSG00000255899 | AL662844.2 | HLA-B associated transcript 1 (BAT1) pseudogene | |
| ENSG00000204525 | HLA-C | major histocompatibility complex, class I, C [Source:HGNC Symbol;Acc:HGNC:4933] | Precursor B-cell acute lymphoblastic leukemia |
| ENSG00000204525 | HLA-C | major histocompatibility complex, class I, C [Source:HGNC Symbol;Acc:HGNC:4933] | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 SUSCEPTIBILITY TO |
| ENSG00000204525 | HLA-C | major histocompatibility complex, class I, C [Source:HGNC Symbol;Acc:HGNC:4933] | PSORIASIS 1 SUSCEPTIBILITY TO |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Abacavir toxicity |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Allopurinol toxicity |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Behcet disease |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Flucloxacilline toxicity |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Phenytoin or carbamazepine toxicity |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Pulmonary arterial hypertension associated with connective tissue disease |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Stevens-Johnson syndrome |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | Takayasu arteritis |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | SEVERE CUTANEOUS ADVERSE REACTION SUSCEPTIBILITY TO TOXIC EPIDERMAL NECROLYSIS SUSCEPTIBILITY TO INCLUDED |
| ENSG00000234745 | HLA-B | major histocompatibility complex, class I, B [Source:HGNC Symbol;Acc:HGNC:4932] | SPONDYLOARTHROPATHY SUSCEPTIBILITY TO 1 |
| ENSG00000214892 | USP8P1 | ubiquitin specific peptidase 8 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:13987] | |
| ENSG00000227939 | RPL3P2 | ribosomal protein L3 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:17834] | |
| ENSG00000231402 | WASF5P | WASP family member 5, pseudogene [Source:HGNC Symbol;Acc:HGNC:21665] | |
| ENSG00000256166 | LINC02571 | long intergenic non-protein coding RNA 2571 [Source:HGNC Symbol;Acc:HGNC:53630] | |
| ENSG00000229836 | AL671883.1 | novel HLA class I pseudogene fragment | |
| ENSG00000277402 | MIR6891 | microRNA 6891 [Source:HGNC Symbol;Acc:HGNC:50243] | |
| ENSG00000271581 | AL671883.2 | HLA complex group 4 (HCG4) pseudogene | |
| ENSG00000228432 | DHFRP2 | dihydrofolate reductase pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:2863] | |
| ENSG00000285647 | AL671883.3 | novel transcript | |
| ENSG00000201658 | RNU6-283P | RNA, U6 small nuclear 283, pseudogene [Source:HGNC Symbol;Acc:HGNC:47246] | |
| ENSG00000230994 | FGFR3P1 | fibroblast growth factor receptor 3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21664] | |
| ENSG00000223702 | ZDHHC20P2 | zinc finger DHHC-type containing 20 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:33457] | |
| ENSG00000225851 | HLA-S | major histocompatibility complex, class I, S (pseudogene) [Source:HGNC Symbol;Acc:HGNC:19395] | |
| ENSG00000272221 | AL645933.3 | novel transcript | |
| ENSG00000204520 | MICA | MHC class I polypeptide-related sequence A [Source:HGNC Symbol;Acc:HGNC:7090] | |
| ENSG00000288587 | AL645933.5 | novel transcript | |
| ENSG00000199332 | AL645933.1 | ||
| ENSG00000230174 | LINC01149 | long intergenic non-protein coding RNA 1149 [Source:HGNC Symbol;Acc:HGNC:39757] | |
| ENSG00000233902 | AL645933.2 | HLA complex group 4 (HCG4) pseudogene | |
| ENSG00000206337 | HCP5 | HLA complex P5 [Source:HGNC Symbol;Acc:HGNC:21659] | |
| ENSG00000286940 | AL645933.4 | novel transcript | |
| ENSG00000204516 | MICB | MHC class I polypeptide-related sequence B [Source:HGNC Symbol;Acc:HGNC:7091] | |
| ENSG00000201680 | AL663061.1 | ||
| ENSG00000256851 | AL663061.2 | HLA pseudogene | |
| ENSG00000219797 | PPIAP9 | peptidylprolyl isomerase A pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:9272] | |
| ENSG00000225499 | RPL15P4 | ribosomal protein L15 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:21663] | |
| ENSG00000204511 | MCCD1 | mitochondrial coiled-coil domain 1 [Source:HGNC Symbol;Acc:HGNC:20668] | |
| ENSG00000198563 | DDX39B | DExD-box helicase 39B [Source:HGNC Symbol;Acc:HGNC:13917] | |
| ENSG00000254870 | ATP6V1G2-DDX39B | ATP6V1G2-DDX39B readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:41999] | |
| ENSG00000201785 | SNORD117 | small nucleolar RNA, C/D box 117 [Source:HGNC Symbol;Acc:HGNC:32742] | |
| ENSG00000265236 | SNORD84 | small nucleolar RNA, C/D box 84 [Source:HGNC Symbol;Acc:HGNC:32743] | |
| ENSG00000234006 | DDX39B-AS1 | DDX39B antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39771] | |
| ENSG00000213760 | ATP6V1G2 | ATPase H+ transporting V1 subunit G2 [Source:HGNC Symbol;Acc:HGNC:862] | |
| ENSG00000204498 | NFKBIL1 | NFKB inhibitor like 1 [Source:HGNC Symbol;Acc:HGNC:7800] | Rheumatoid arthritis |
| ENSG00000226979 | LTA | lymphotoxin alpha [Source:HGNC Symbol;Acc:HGNC:6709] | LEPROSY SUSCEPTIBILITY TO 4 |
| ENSG00000226979 | LTA | lymphotoxin alpha [Source:HGNC Symbol;Acc:HGNC:6709] | MYOCARDIAL INFARCTION SUSCEPTIBILITY TO MYOCARDIAL INFARCTION SUSCEPTIBILITY TO 1 INCLUDED |
| ENSG00000226979 | LTA | lymphotoxin alpha [Source:HGNC Symbol;Acc:HGNC:6709] | PSORIATIC ARTHRITIS SUSCEPTIBILITY TOPSORIATIC ARTHRITIS SUSCEPTIBILITY TO 1 INCLUDED |
| ENSG00000232810 | TNF | tumor necrosis factor [Source:HGNC Symbol;Acc:HGNC:11892] | ASTHMA SUSCEPTIBILITY TO |
| ENSG00000232810 | TNF | tumor necrosis factor [Source:HGNC Symbol;Acc:HGNC:11892] | MALARIA SUSCEPTIBILITY TO MALARIA RESISTANCE TO INCLUDED |
| ENSG00000232810 | TNF | tumor necrosis factor [Source:HGNC Symbol;Acc:HGNC:11892] | MIGRAINE WITH OR WITHOUT AURA SUSCEPTIBILITY TO 1 |
| ENSG00000227507 | LTB | lymphotoxin beta [Source:HGNC Symbol;Acc:HGNC:6711] | |
| ENSG00000204482 | LST1 | leukocyte specific transcript 1 [Source:HGNC Symbol;Acc:HGNC:14189] | |
| ENSG00000204475 | NCR3 | natural cytotoxicity triggering receptor 3 [Source:HGNC Symbol;Acc:HGNC:19077] | MALARIA MILD SUSCEPTIBILITY TO |
| ENSG00000230622 | UQCRHP1 | ubiquinol-cytochrome c reductase hinge protein pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31341] | |
| ENSG00000204472 | AIF1 | allograft inflammatory factor 1 [Source:HGNC Symbol;Acc:HGNC:352] | |
| ENSG00000204469 | PRRC2A | proline rich coiled-coil 2A [Source:HGNC Symbol;Acc:HGNC:13918] | |
| ENSG00000200816 | SNORA38 | small nucleolar RNA, H/ACA box 38 [Source:HGNC Symbol;Acc:HGNC:32631] | |
| ENSG00000274494 | MIR6832 | microRNA 6832 [Source:HGNC Symbol;Acc:HGNC:50140] | |
| ENSG00000204463 | BAG6 | BCL2 associated athanogene 6 [Source:HGNC Symbol;Acc:HGNC:13919] | |
| ENSG00000204444 | APOM | apolipoprotein M [Source:HGNC Symbol;Acc:HGNC:13916] | |
| ENSG00000204439 | C6orf47 | chromosome 6 open reading frame 47 [Source:HGNC Symbol;Acc:HGNC:19076] | |
| ENSG00000227198 | C6orf47-AS1 | C6orf47 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39767] | |
| ENSG00000204438 | GPANK1 | G-patch domain and ankyrin repeats 1 [Source:HGNC Symbol;Acc:HGNC:13920] | |
| ENSG00000201207 | AL662899.1 | ||
| ENSG00000204435 | CSNK2B | casein kinase 2 beta [Source:HGNC Symbol;Acc:HGNC:2460] | Autosomal dominant non-syndromic intellectual disability |
| ENSG00000263020 | AL662899.4 | novel protein | |
| ENSG00000240053 | LY6G5B | lymphocyte antigen 6 family member G5B [Source:HGNC Symbol;Acc:HGNC:13931] | |
| ENSG00000204428 | LY6G5C | lymphocyte antigen 6 family member G5C [Source:HGNC Symbol;Acc:HGNC:13932] | |
| ENSG00000204427 | ABHD16A | abhydrolase domain containing 16A [Source:HGNC Symbol;Acc:HGNC:13921] | |
| ENSG00000204422 | AL662899.3 | novel transcript | |
| ENSG00000266776 | MIR4646 | microRNA 4646 [Source:HGNC Symbol;Acc:HGNC:41543] | |
| ENSG00000204424 | LY6G6F | lymphocyte antigen 6 family member G6F [Source:HGNC Symbol;Acc:HGNC:13933] | |
| ENSG00000250641 | LY6G6F-LY6G6D | LY6G6F-LY6G6D readthrough [Source:NCBI gene;Acc:110599563] | |
| ENSG00000255552 | LY6G6E | lymphocyte antigen 6 family member G6E [Source:HGNC Symbol;Acc:HGNC:13934] | |
| ENSG00000244355 | LY6G6D | lymphocyte antigen 6 family member G6D [Source:HGNC Symbol;Acc:HGNC:13935] | |
| ENSG00000204420 | MPIG6B | megakaryocyte and platelet inhibitory receptor G6b [Source:HGNC Symbol;Acc:HGNC:13937] | THROMBOCYTOPENIA ANEMIA AND MYELOFIBROSIS |
| ENSG00000204421 | LY6G6C | lymphocyte antigen 6 family member G6C [Source:HGNC Symbol;Acc:HGNC:13936] | |
| ENSG00000213722 | DDAH2 | dimethylarginine dimethylaminohydrolase 2 [Source:HGNC Symbol;Acc:HGNC:2716] | |
| ENSG00000213719 | CLIC1 | chloride intracellular channel 1 [Source:HGNC Symbol;Acc:HGNC:2062] | |
| ENSG00000204410 | MSH5 | mutS homolog 5 [Source:HGNC Symbol;Acc:HGNC:7328] | PREMATURE OVARIAN FAILURE 13 |
| ENSG00000255152 | MSH5-SAPCD1 | MSH5-SAPCD1 readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:41994] | |
| ENSG00000252743 | RNU6-850P | RNA, U6 small nuclear 850, pseudogene [Source:HGNC Symbol;Acc:HGNC:47813] | |
| ENSG00000228727 | SAPCD1 | suppressor APC domain containing 1 [Source:HGNC Symbol;Acc:HGNC:13938] | |
| ENSG00000235663 | SAPCD1-AS1 | SAPCD1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39824] | |
| ENSG00000204396 | VWA7 | von Willebrand factor A domain containing 7 [Source:HGNC Symbol;Acc:HGNC:13939] | |
| ENSG00000204394 | VARS1 | valyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:12651] | NEURODEVELOPMENTAL DISORDER WITH MICROCEPHALY SEIZURES AND CORTICAL ATROPHY |
| ENSG00000201555 | AL662899.2 | ||
| ENSG00000204392 | LSM2 | LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated [Source:HGNC Symbol;Acc:HGNC:13940] | |
| ENSG00000204390 | HSPA1L | heat shock protein family A (Hsp70) member 1 like [Source:HGNC Symbol;Acc:HGNC:5234] | |
| ENSG00000204389 | HSPA1A | heat shock protein family A (Hsp70) member 1A [Source:HGNC Symbol;Acc:HGNC:5232] | |
| ENSG00000204388 | HSPA1B | heat shock protein family A (Hsp70) member 1B [Source:HGNC Symbol;Acc:HGNC:5233] | |
| ENSG00000285565 | AL671762.1 | novel transcript | |
| ENSG00000204387 | SNHG32 | small nucleolar RNA host gene 32 [Source:HGNC Symbol;Acc:HGNC:19078] | |
| ENSG00000201823 | SNORD48 | small nucleolar RNA, C/D box 48 [Source:HGNC Symbol;Acc:HGNC:10188] | |
| ENSG00000201754 | SNORD52 | small nucleolar RNA, C/D box 52 [Source:HGNC Symbol;Acc:HGNC:10202] | |
| ENSG00000204386 | NEU1 | neuraminidase 1 [Source:HGNC Symbol;Acc:HGNC:7758] | Congenital sialidosis type 2 |
| ENSG00000204386 | NEU1 | neuraminidase 1 [Source:HGNC Symbol;Acc:HGNC:7758] | Juvenile sialidosis type 2 |
| ENSG00000204386 | NEU1 | neuraminidase 1 [Source:HGNC Symbol;Acc:HGNC:7758] | Sialidosis type 1 |
| ENSG00000204386 | NEU1 | neuraminidase 1 [Source:HGNC Symbol;Acc:HGNC:7758] | Sialidosis |
| ENSG00000204386 | NEU1 | neuraminidase 1 [Source:HGNC Symbol;Acc:HGNC:7758] | NEURAMINIDASE DEFICIENCY |
| ENSG00000204385 | SLC44A4 | solute carrier family 44 member 4 [Source:HGNC Symbol;Acc:HGNC:13941] | Autosomal dominant non-syndromic sensorineural deafness type DFNA |
| ENSG00000204385 | SLC44A4 | solute carrier family 44 member 4 [Source:HGNC Symbol;Acc:HGNC:13941] | DEAFNESS AUTOSOMAL DOMINANT 72 |
| ENSG00000237080 | EHMT2-AS1 | EHMT2 and SLC44A4 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39751] | |
| ENSG00000204371 | EHMT2 | euchromatic histone lysine methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:14129] | |
| ENSG00000166278 | C2 | complement C2 [Source:HGNC Symbol;Acc:HGNC:1248] | Immunodeficiency due to a classical component pathway complement deficiency |
| ENSG00000166278 | C2 | complement C2 [Source:HGNC Symbol;Acc:HGNC:1248] | COMPLEMENT COMPONENT 2 DEFICIENCY |
| ENSG00000166278 | C2 | complement C2 [Source:HGNC Symbol;Acc:HGNC:1248] | MACULAR DEGENERATION AGE-RELATED 14 |
| ENSG00000204366 | ZBTB12 | zinc finger and BTB domain containing 12 [Source:HGNC Symbol;Acc:HGNC:19066] | |
| ENSG00000244255 | AL645922.1 | novel complement component 2 (C2) and complement factor B (CFB) protein | |
| ENSG00000281756 | C2-AS1 | C2 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:49464] | |
| ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] | Atypical hemolytic uremic syndrome with complement gene abnormality |
| ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] | COMPLEMENT FACTOR B DEFICIENCY |
| ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] | HEMOLYTIC UREMIC SYNDROME ATYPICAL SUSCEPTIBILITY TO 4 |
| ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] | MACULAR DEGENERATION AGE-RELATED 14 |
| ENSG00000204356 | NELFE | negative elongation factor complex member E [Source:HGNC Symbol;Acc:HGNC:13974] | |
| ENSG00000284446 | MIR1236 | microRNA 1236 [Source:HGNC Symbol;Acc:HGNC:33925] | |
| ENSG00000204351 | SKIV2L | Ski2 like RNA helicase [Source:HGNC Symbol;Acc:HGNC:10898] | Syndromic diarrhea |
| ENSG00000204351 | SKIV2L | Ski2 like RNA helicase [Source:HGNC Symbol;Acc:HGNC:10898] | TRICHOHEPATOENTERIC SYNDROME 2 |
| ENSG00000204348 | DXO | decapping exoribonuclease [Source:HGNC Symbol;Acc:HGNC:2992] | |
| ENSG00000204344 | STK19 | serine/threonine kinase 19 [Source:HGNC Symbol;Acc:HGNC:11398] | |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | Autosomal systemic lupus erythematosus |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | Behcet disease |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | Immunodeficiency due to a classical component pathway complement deficiency |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | Systemic lupus erythematosus |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | BLOOD GROUP CHIDO/RODGERS SYSTEM |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) [Source:HGNC Symbol;Acc:HGNC:1323] | COMPLEMENT COMPONENT 4A DEFICIENCY |
| ENSG00000233627 | C4A-AS1 | C4A antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39753] | |
| ENSG00000204338 | CYP21A1P | cytochrome P450 family 21 subfamily A member 1, pseudogene [Source:HGNC Symbol;Acc:HGNC:2599] | |
| ENSG00000248290 | TNXA | tenascin XA (pseudogene) [Source:HGNC Symbol;Acc:HGNC:11975] | |
| ENSG00000250535 | STK19B | serine/threonine kinase 19B (pseudogene) [Source:HGNC Symbol;Acc:HGNC:21668] | |
| ENSG00000224389 | C4B | complement C4B (Chido blood group) [Source:HGNC Symbol;Acc:HGNC:1324] | Immunodeficiency due to a classical component pathway complement deficiency |
| ENSG00000224389 | C4B | complement C4B (Chido blood group) [Source:HGNC Symbol;Acc:HGNC:1324] | Systemic lupus erythematosus |
| ENSG00000224389 | C4B | complement C4B (Chido blood group) [Source:HGNC Symbol;Acc:HGNC:1324] | COMPLEMENT COMPONENT 4B DEFICIENCY |
| ENSG00000229776 | C4B-AS1 | C4B antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39752] | |
| ENSG00000231852 | CYP21A2 | cytochrome P450 family 21 subfamily A member 2 [Source:HGNC Symbol;Acc:HGNC:2600] | Classic congenital adrenal hyperplasia due to 21-hydroxylase deficiency salt wasting form |
| ENSG00000231852 | CYP21A2 | cytochrome P450 family 21 subfamily A member 2 [Source:HGNC Symbol;Acc:HGNC:2600] | Classic congenital adrenal hyperplasia due to 21-hydroxylase deficiency simple virilizing form |
| ENSG00000231852 | CYP21A2 | cytochrome P450 family 21 subfamily A member 2 [Source:HGNC Symbol;Acc:HGNC:2600] | ADRENAL HYPERPLASIA CONGENITAL DUE TO 21-HYDROXYLASE DEFICIENCY |
| ENSG00000168477 | TNXB | tenascin XB [Source:HGNC Symbol;Acc:HGNC:11976] | Classical-like Ehlers-Danlos syndrome type 1 |
| ENSG00000168477 | TNXB | tenascin XB [Source:HGNC Symbol;Acc:HGNC:11976] | Familial vesicoureteral reflux |
| ENSG00000168477 | TNXB | tenascin XB [Source:HGNC Symbol;Acc:HGNC:11976] | EHLERS-DANLOS SYNDROME CLASSIC-LIKE |
| ENSG00000168477 | TNXB | tenascin XB [Source:HGNC Symbol;Acc:HGNC:11976] | Vesicoureteral reflux 8 |
| ENSG00000252512 | RNA5SP206 | RNA, 5S ribosomal pseudogene 206 [Source:HGNC Symbol;Acc:HGNC:43106] | |
| ENSG00000284829 | AL662884.2 | novel transcript | |
| ENSG00000286974 | AL662884.5 | novel transcript, antisense to TNXB | |
| ENSG00000213676 | ATF6B | activating transcription factor 6 beta [Source:HGNC Symbol;Acc:HGNC:2349] | |
| ENSG00000204315 | FKBPL | FKBP prolyl isomerase like [Source:HGNC Symbol;Acc:HGNC:13949] | |
| ENSG00000204314 | PRRT1 | proline rich transmembrane protein 1 [Source:HGNC Symbol;Acc:HGNC:13943] | |
| ENSG00000285085 | AL662884.4 | novel protein | |
| ENSG00000284954 | AL662884.3 | novel transcript | |
| ENSG00000221988 | PPT2 | palmitoyl-protein thioesterase 2 [Source:HGNC Symbol;Acc:HGNC:9326] | |
| ENSG00000258388 | PPT2-EGFL8 | PPT2-EGFL8 readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:48343] | |
| ENSG00000241404 | EGFL8 | EGF like domain multiple 8 [Source:HGNC Symbol;Acc:HGNC:13944] | |
| ENSG00000204310 | AGPAT1 | 1-acylglycerol-3-phosphate O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:324] | |
| ENSG00000284469 | MIR6721 | microRNA 6721 [Source:HGNC Symbol;Acc:HGNC:50049] | |
| ENSG00000204308 | RNF5 | ring finger protein 5 [Source:HGNC Symbol;Acc:HGNC:10068] | |
| ENSG00000277264 | MIR6833 | microRNA 6833 [Source:HGNC Symbol;Acc:HGNC:50245] | |
| ENSG00000204305 | AGER | advanced glycosylation end-product specific receptor [Source:HGNC Symbol;Acc:HGNC:320] | |
| ENSG00000273333 | AL662884.1 | novel transcript | |
| ENSG00000204304 | PBX2 | PBX homeobox 2 [Source:HGNC Symbol;Acc:HGNC:8633] | |
| ENSG00000213654 | GPSM3 | G protein signaling modulator 3 [Source:HGNC Symbol;Acc:HGNC:13945] | |
| ENSG00000204301 | NOTCH4 | notch receptor 4 [Source:HGNC Symbol;Acc:HGNC:7884] | |
| ENSG00000225914 | TSBP1-AS1 | TSBP1 and BTNL2 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39756] | |
| ENSG00000204296 | TSBP1 | testis expressed basic protein 1 [Source:HGNC Symbol;Acc:HGNC:13922] | |
| ENSG00000237285 | HNRNPA1P2 | heterogeneous nuclear ribonucleoprotein A1 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:13958] | |
| ENSG00000223335 | RNU6-603P | RNA, U6 small nuclear 603, pseudogene [Source:HGNC Symbol;Acc:HGNC:47566] | |
| ENSG00000204290 | BTNL2 | butyrophilin like 2 [Source:HGNC Symbol;Acc:HGNC:1142] | Sarcoidosis |
| ENSG00000204290 | BTNL2 | butyrophilin like 2 [Source:HGNC Symbol;Acc:HGNC:1142] | SARCOIDOSIS SUSCEPTIBILITY TO 2 |
| ENSG00000204287 | HLA-DRA | major histocompatibility complex, class II, DR alpha [Source:HGNC Symbol;Acc:HGNC:4947] | Graham Little-Piccardi-Lassueur syndrome |
| ENSG00000196301 | HLA-DRB9 | major histocompatibility complex, class II, DR beta 9 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4957] | |
| ENSG00000198502 | HLA-DRB5 | major histocompatibility complex, class II, DR beta 5 [Source:HGNC Symbol;Acc:HGNC:4953] | |
| ENSG00000251916 | RNU1-61P | RNA, U1 small nuclear 61, pseudogene [Source:HGNC Symbol;Acc:HGNC:48403] | |
| ENSG00000229391 | HLA-DRB6 | major histocompatibility complex, class II, DR beta 6 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4954] | |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Bullous pemphigoid |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Diffuse cutaneous systemic sclerosis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Follicular lymphoma |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Limbic encephalitis with LGI1 antibodies |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Limited cutaneous systemic sclerosis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Limited systemic sclerosis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Narcolepsy type 1 |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Narcolepsy type 2 |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Pediatric multiple sclerosis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Sarcoidosis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Systemic lupus erythematosus |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | Systemic-onset juvenile idiopathic arthritis |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | MULTIPLE SCLEROSIS SUSCEPTIBILITY TO |
| ENSG00000196126 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 [Source:HGNC Symbol;Acc:HGNC:4948] | SARCOIDOSIS SUSCEPTIBILITY TO 1 |
| ENSG00000196735 | HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 [Source:HGNC Symbol;Acc:HGNC:4942] | Adult-onset myasthenia gravis |
| ENSG00000196735 | HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 [Source:HGNC Symbol;Acc:HGNC:4942] | Idiopathic achalasia |
| ENSG00000196735 | HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 [Source:HGNC Symbol;Acc:HGNC:4942] | CELIAC DISEASE SUSCEPTIBILITY TO 1 |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Bullous pemphigoid |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Idiopathic achalasia |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Limbic encephalitis with LGI1 antibodies |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Narcolepsy type 1 |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Narcolepsy type 2 |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | Pediatric multiple sclerosis |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | CELIAC DISEASE SUSCEPTIBILITY TO 1 |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | CREUTZFELDT-JAKOB DISEASE |
| ENSG00000179344 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 [Source:HGNC Symbol;Acc:HGNC:4944] | MULTIPLE SCLEROSIS SUSCEPTIBILITY TO |
| ENSG00000223534 | HLA-DQB1-AS1 | HLA-DQB1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39762] | |
| ENSG00000235040 | MTCO3P1 | MT-CO3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31342] | |
| ENSG00000232080 | AL662789.1 | novel transcript | |
| ENSG00000226030 | HLA-DQB3 | major histocompatibility complex, class II, DQ beta 3 [Source:HGNC Symbol;Acc:HGNC:4946] | |
| ENSG00000237541 | HLA-DQA2 | major histocompatibility complex, class II, DQ alpha 2 [Source:HGNC Symbol;Acc:HGNC:4943] | CELIAC DISEASE SUSCEPTIBILITY TO 1 |
| ENSG00000263649 | MIR3135B | microRNA 3135b [Source:HGNC Symbol;Acc:HGNC:41783] | |
| ENSG00000232629 | HLA-DQB2 | major histocompatibility complex, class II, DQ beta 2 [Source:HGNC Symbol;Acc:HGNC:4945] | |
| ENSG00000241106 | HLA-DOB | major histocompatibility complex, class II, DO beta [Source:HGNC Symbol;Acc:HGNC:4937] | |
| ENSG00000250264 | AL669918.1 | novel protein, TAP2-HLA-DOB readthrough | |
| ENSG00000204267 | TAP2 | transporter 2, ATP binding cassette subfamily B member [Source:HGNC Symbol;Acc:HGNC:44] | Immunodeficiency by defective expression of MHC class I |
| ENSG00000204267 | TAP2 | transporter 2, ATP binding cassette subfamily B member [Source:HGNC Symbol;Acc:HGNC:44] | BARE LYMPHOCYTE SYNDROME TYPE I |
| ENSG00000204264 | PSMB8 | proteasome 20S subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] | CANDLE syndrome |
| ENSG00000204264 | PSMB8 | proteasome 20S subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] | JMP syndrome |
| ENSG00000204264 | PSMB8 | proteasome 20S subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] | Nakajo-Nishimura syndrome |
| ENSG00000204264 | PSMB8 | proteasome 20S subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] | Nakajo syndrome |
| ENSG00000204264 | PSMB8 | proteasome 20S subunit beta 8 [Source:HGNC Symbol;Acc:HGNC:9545] | PROTEASOME-ASSOCIATED AUTOINFLAMMATORY SYNDROME 1 |
| ENSG00000204261 | PSMB8-AS1 | PSMB8 antisense RNA 1 (head to head) [Source:HGNC Symbol;Acc:HGNC:39758] | |
| ENSG00000240065 | PSMB9 | proteasome 20S subunit beta 9 [Source:HGNC Symbol;Acc:HGNC:9546] | PROTEASOME-ASSOCIATED AUTOINFLAMMATORY SYNDROME 3 |
| ENSG00000168394 | TAP1 | transporter 1, ATP binding cassette subfamily B member [Source:HGNC Symbol;Acc:HGNC:43] | Immunodeficiency by defective expression of MHC class I |
| ENSG00000168394 | TAP1 | transporter 1, ATP binding cassette subfamily B member [Source:HGNC Symbol;Acc:HGNC:43] | BARE LYMPHOCYTE SYNDROME TYPE I |
| ENSG00000234515 | PPP1R2P1 | protein phosphatase 1 regulatory inhibitor subunit 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:9289] | |
| ENSG00000235301 | HLA-Z | major histocompatibility complex, class I, Z (pseudogene) [Source:HGNC Symbol;Acc:HGNC:19394] | |
| ENSG00000242574 | HLA-DMB | major histocompatibility complex, class II, DM beta [Source:HGNC Symbol;Acc:HGNC:4935] | |
| ENSG00000248993 | AL645941.2 | novel protein | |
| ENSG00000204257 | HLA-DMA | major histocompatibility complex, class II, DM alpha [Source:HGNC Symbol;Acc:HGNC:4934] | |
| ENSG00000204256 | BRD2 | bromodomain containing 2 [Source:HGNC Symbol;Acc:HGNC:1103] | |
| ENSG00000223837 | AL645941.1 | novel transcript | |
| ENSG00000263756 | AL645941.3 | novel transcript, antisense to BRD2 | |
| ENSG00000204252 | HLA-DOA | major histocompatibility complex, class II, DO alpha [Source:HGNC Symbol;Acc:HGNC:4936] | |
| ENSG00000231389 | HLA-DPA1 | major histocompatibility complex, class II, DP alpha 1 [Source:HGNC Symbol;Acc:HGNC:4938] | Granulomatosis with polyangiitis |
| ENSG00000223865 | HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 [Source:HGNC Symbol;Acc:HGNC:4940] | Chronic beryllium disease |
| ENSG00000223865 | HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 [Source:HGNC Symbol;Acc:HGNC:4940] | Granulomatosis with polyangiitis |
| ENSG00000224796 | RPL32P1 | ribosomal protein L32 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:10339] | |
| ENSG00000231461 | HLA-DPA2 | major histocompatibility complex, class II, DP alpha 2 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4939] | |
| ENSG00000228688 | COL11A2P1 | collagen type XI alpha 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:13947] | |
| ENSG00000224557 | HLA-DPB2 | major histocompatibility complex, class II, DP beta 2 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:4941] | |
| ENSG00000237398 | HLA-DPA3 | major histocompatibility complex, class II, DP alpha 3 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:19393] | |
| ENSG00000230313 | HCG24 | HLA complex group 24 [Source:HGNC Symbol;Acc:HGNC:23500] | |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | Autosomal dominant non-syndromic sensorineural deafness type DFNA |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | Autosomal recessive non-syndromic sensorineural deafness type DFNB |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | Fibrochondrogenesis |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | Otospondylomegaepiphyseal dysplasia |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | STICKLER SYNDROME TYPE 3 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | Weissenbacher-Zweymuller syndrome |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | AUTOSOMAL RECESSIVE OTOSPONDYLOMEGAEPIPHYSEAL DYSPLASIA |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | DEAFNESS AUTOSOMAL DOMINANT TYPE 13 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | DEAFNESS AUTOSOMAL RECESSIVE TYPE 53 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | WEISSENBACHER-ZWEYMUELLER SYNDROME |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | DEAFNESS AUTOSOMAL DOMINANT 13 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | DEAFNESS AUTOSOMAL RECESSIVE 53 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | FIBROCHONDROGENESIS 2 |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | OTOSPONDYLOMEGAEPIPHYSEAL DYSPLASIA AUTOSOMAL DOMINANT |
| ENSG00000204248 | COL11A2 | collagen type XI alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2187] | OTOSPONDYLOMEGAEPIPHYSEAL DYSPLASIA AUTOSOMAL RECESSIVE |
| ENSG00000204231 | RXRB | retinoid X receptor beta [Source:HGNC Symbol;Acc:HGNC:10478] | |
| ENSG00000202441 | RNY4P10 | RNY4 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:34060] | |
| ENSG00000112473 | SLC39A7 | solute carrier family 39 member 7 [Source:HGNC Symbol;Acc:HGNC:4927] | |
| ENSG00000204228 | HSD17B8 | hydroxysteroid 17-beta dehydrogenase 8 [Source:HGNC Symbol;Acc:HGNC:3554] | |
| ENSG00000199036 | MIR219A1 | microRNA 219a-1 [Source:HGNC Symbol;Acc:HGNC:31597] | |
| ENSG00000204227 | RING1 | ring finger protein 1 [Source:HGNC Symbol;Acc:HGNC:10018] | |
| ENSG00000225463 | ZNF70P1 | zinc finger protein 70 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:13846] | |
| ENSG00000223457 | HTATSF1P1 | HIV-1 Tat specific factor 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:13959] | |
| ENSG00000272217 | AL645940.1 | novel transcript | |
| ENSG00000232940 | HCG25 | HLA complex group 25 [Source:HGNC Symbol;Acc:HGNC:20196] | |
| ENSG00000223501 | VPS52 | VPS52 subunit of GARP complex [Source:HGNC Symbol;Acc:HGNC:10518] | |
| ENSG00000231500 | RPS18 | ribosomal protein S18 [Source:HGNC Symbol;Acc:HGNC:10401] | |
| ENSG00000235863 | B3GALT4 | beta-1,3-galactosyltransferase 4 [Source:HGNC Symbol;Acc:HGNC:919] | |
| ENSG00000227057 | WDR46 | WD repeat domain 46 [Source:HGNC Symbol;Acc:HGNC:13923] | |
| ENSG00000284517 | MIR6873 | microRNA 6873 [Source:HGNC Symbol;Acc:HGNC:50231] | |
| ENSG00000204220 | PFDN6 | prefoldin subunit 6 [Source:HGNC Symbol;Acc:HGNC:4926] | |
| ENSG00000284064 | MIR6834 | microRNA 6834 [Source:HGNC Symbol;Acc:HGNC:50108] | |
| ENSG00000237441 | RGL2 | ral guanine nucleotide dissociation stimulator like 2 [Source:HGNC Symbol;Acc:HGNC:9769] | |
| ENSG00000231925 | TAPBP | TAP binding protein [Source:HGNC Symbol;Acc:HGNC:11566] | Immunodeficiency by defective expression of MHC class I |
| ENSG00000231925 | TAPBP | TAP binding protein [Source:HGNC Symbol;Acc:HGNC:11566] | BARE LYMPHOCYTE SYNDROME TYPE I |
| ENSG00000236104 | ZBTB22 | zinc finger and BTB domain containing 22 [Source:HGNC Symbol;Acc:HGNC:13085] | |
| ENSG00000204209 | DAXX | death domain associated protein [Source:HGNC Symbol;Acc:HGNC:2681] | Neuroendocrine tumor of stomach |
| ENSG00000285064 | SMIM40 | small integral membrane protein 40 [Source:HGNC Symbol;Acc:HGNC:54073] | |
| ENSG00000286920 | SMIM40 | small integral membrane protein 40 [Source:NCBI gene;Acc:113523636] | |
| ENSG00000229596 | MYL8P | myosin light chain 8, pseudogene [Source:HGNC Symbol;Acc:HGNC:7589] | |
| ENSG00000228285 | LYPLA2P1 | LYPLA2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21069] | |
| ENSG00000225644 | RPL35AP4 | ribosomal protein L35a pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:33460] | |
| ENSG00000237649 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | |
| ENSG00000204194 | RPL12P1 | ribosomal protein L12 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:13976] | |
| ENSG00000112511 | PHF1 | PHD finger protein 1 [Source:HGNC Symbol;Acc:HGNC:8919] | |
| ENSG00000112514 | CUTA | cutA divalent cation tolerance homolog [Source:HGNC Symbol;Acc:HGNC:21101] | |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | Autosomal dominant non-syndromic intellectual disability |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | SYNGAP1-related developmental and epileptic encephalopathy |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | Undetermined early-onset epileptic encephalopathy |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | Epileptic encephalopathy |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | MENTAL RETARDATION AUTOSOMAL DOMINANT TYPE 5 |
| ENSG00000197283 | SYNGAP1 | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | MENTAL RETARDATION AUTOSOMAL DOMINANT 5 |
| ENSG00000274259 | SYNGAP1-AS1 | SYNGAP1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:53831] | |
| ENSG00000284256 | MIR5004 | microRNA 5004 [Source:HGNC Symbol;Acc:HGNC:43532] | |
| ENSG00000213588 | ZBTB9 | zinc finger and BTB domain containing 9 [Source:HGNC Symbol;Acc:HGNC:28323] | |
| ENSG00000204188 | GGNBP1 | gametogenetin binding protein 1 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:19427] | |
| ENSG00000242014 | RN7SL26P | RNA, 7SL, cytoplasmic 26, pseudogene [Source:HGNC Symbol;Acc:HGNC:46042] | |
| ENSG00000276746 | Z93017.1 | ||
| ENSG00000030110 | BAK1 | BCL2 antagonist/killer 1 [Source:HGNC Symbol;Acc:HGNC:949] | |
| ENSG00000197251 | LINC00336 | long intergenic non-protein coding RNA 336 [Source:HGNC Symbol;Acc:HGNC:33813] | |
| ENSG00000096433 | ITPR3 | inositol 1,4,5-trisphosphate receptor type 3 [Source:HGNC Symbol;Acc:HGNC:6182] | DIABETES MELLITUS INSULIN-DEPENDENT |
| ENSG00000137288 | UQCC2 | ubiquinol-cytochrome c reductase complex assembly factor 2 [Source:HGNC Symbol;Acc:HGNC:21237] | Isolated complex III deficiency |
| ENSG00000137288 | UQCC2 | ubiquinol-cytochrome c reductase complex assembly factor 2 [Source:HGNC Symbol;Acc:HGNC:21237] | MITOCHONDRIAL COMPLEX III DEFICIENCY NUCLEAR TYPE 7 |
| ENSG00000266509 | MIR3934 | microRNA 3934 [Source:HGNC Symbol;Acc:HGNC:38965] | |
| ENSG00000161896 | IP6K3 | inositol hexakisphosphate kinase 3 [Source:HGNC Symbol;Acc:HGNC:17269] | |
| ENSG00000161904 | LEMD2 | LEM domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21244] | Early-onset posterior subcapsular cataract |
| ENSG00000161904 | LEMD2 | LEM domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21244] | Total early-onset cataract |
| ENSG00000161904 | LEMD2 | LEM domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21244] | Nuclear Envelopathy with Early Progeroid Appearance |
| ENSG00000161904 | LEMD2 | LEM domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21244] | CATARACT 46 JUVENILE-ONSET |
| ENSG00000096395 | MLN | motilin [Source:HGNC Symbol;Acc:HGNC:7141] | |
| ENSG00000287089 | AL138889.3 | novel transcript, antisense to MLN | |
| ENSG00000249346 | LINC01016 | long intergenic non-protein coding RNA 1016 [Source:HGNC Symbol;Acc:HGNC:48991] | |
| ENSG00000271362 | AL138889.2 | ankyrin repeat domain 36C (ANKRD36C) pseudogene | |
| ENSG00000233183 | AL138889.1 | novel transcript | |
| ENSG00000276824 | MIR7159 | microRNA 7159 [Source:HGNC Symbol;Acc:HGNC:49978] | |
| ENSG00000221697 | MIR1275 | microRNA 1275 [Source:HGNC Symbol;Acc:HGNC:35346] | |
| ENSG00000124493 | GRM4 | glutamate metabotropic receptor 4 [Source:HGNC Symbol;Acc:HGNC:4596] | |
| ENSG00000271231 | KRT18P9 | keratin 18 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:17767] | |
| ENSG00000214810 | CYCSP55 | CYCS pseudogene 55 [Source:HGNC Symbol;Acc:HGNC:21252] | |
| ENSG00000137309 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | DIABETES MELLITUS NONINSULIN-DEPENDENT |
| ENSG00000276404 | MIR6835 | microRNA 6835 [Source:HGNC Symbol;Acc:HGNC:49963] | |
| ENSG00000186577 | SMIM29 | small integral membrane protein 29 [Source:HGNC Symbol;Acc:HGNC:1340] | |
| ENSG00000225339 | AL354740.1 | novel transcript, antisense to C6orf1 | |
| ENSG00000220583 | RPL35P2 | ribosomal protein L35 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:35789] | |
| ENSG00000272325 | NUDT3 | nudix hydrolase 3 [Source:HGNC Symbol;Acc:HGNC:8050] | |
| ENSG00000270800 | RPS10-NUDT3 | RPS10-NUDT3 readthrough [Source:HGNC Symbol;Acc:HGNC:49181] | |
| ENSG00000124614 | RPS10 | ribosomal protein S10 [Source:HGNC Symbol;Acc:HGNC:10383] | Blackfan-Diamond anemia |
| ENSG00000124614 | RPS10 | ribosomal protein S10 [Source:HGNC Symbol;Acc:HGNC:10383] | DIAMOND-BLACKFAN ANEMIA 9 |
| ENSG00000124507 | PACSIN1 | protein kinase C and casein kinase substrate in neurons 1 [Source:HGNC Symbol;Acc:HGNC:8570] | |
| ENSG00000124664 | SPDEF | SAM pointed domain containing ETS transcription factor [Source:HGNC Symbol;Acc:HGNC:17257] | |
| ENSG00000196114 | AL031577.1 | interferon induced transmembrane protein (IFITM) pseudogene | |
| ENSG00000196821 | ILRUN | inflammation and lipid regulator with UBA-like and NBR1-like domains [Source:HGNC Symbol;Acc:HGNC:21215] | |
| ENSG00000216636 | RPL7P25 | ribosomal protein L7 pseudogene 25 [Source:HGNC Symbol;Acc:HGNC:36455] | |
| ENSG00000265123 | RN7SL200P | RNA, 7SL, cytoplasmic 200, pseudogene [Source:HGNC Symbol;Acc:HGNC:46216] | |
| ENSG00000220643 | AL031577.2 | ribosomal protein L44 (RPL44) pseudogene | |
| ENSG00000272288 | AL451165.2 | novel transcript, antisense to C6orf106 | |
| ENSG00000186328 | AL451165.1 | ATPase, H+ transporting, lysosomal (vacuolar proton pump) 14kD (ATP6S14) pseudogene | |
| ENSG00000217130 | AL139100.2 | ribosomal protein S10 (RPS10) pseudogene | |
| ENSG00000124562 | SNRPC | small nuclear ribonucleoprotein polypeptide C [Source:HGNC Symbol;Acc:HGNC:11157] | |
| ENSG00000065060 | UHRF1BP1 | UHRF1 binding protein 1 [Source:HGNC Symbol;Acc:HGNC:21216] | |
| ENSG00000206717 | AL139100.1 | ||
| ENSG00000252106 | RNY3P15 | RNY3 pseudogene 15 [Source:HGNC Symbol;Acc:HGNC:50890] | |
| ENSG00000064995 | TAF11 | TATA-box binding protein associated factor 11 [Source:HGNC Symbol;Acc:HGNC:11544] | |
| ENSG00000064999 | ANKS1A | ankyrin repeat and sterile alpha motif domain containing 1A [Source:HGNC Symbol;Acc:HGNC:20961] |
# Or a even nicer table...
# library("DT")
# genes %>% datatableConclusion
See, now we know what is sitting in the region. Any guesses why did 23andMe put a lot of markers there?


Share this post
Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Pinterest
Email